Writing a FASTA file. From Bio import SeqIO for record in SeqIOparseexamplefasta fasta.

Quantitative Extraction Specified Sequence From The Fasta File Python Script Programmer Sought
Is the requested file format.
How to write a fasta file in python. 24112020 from Bio import SeqIO genes fasta_file sysargv1 for entry in SeqIOparse fasta_file fasta. In the case of proteins the amino acids are represented using their one letter acronyms eg. With openunique_genesfasta w as out.
From BioSeqRecord import SeqRecord. Fasta files you might want to look into installing BioPython. A single sequence in FASTA format looks like this.
Filefreadlines fclose return file fasta_fileread_fastaSELEX_100_readstxt printfasta_file The output of fasta file. Genes entryid entryseq Then you can write a new unique fasta file. Here is the code below.
Line linestrip if not line. For record in SeqIOparse fasta fasta. Reading a large FASTA File Very Quickly.
Write a program that will split the genomic DNA into coding and non-coding parts and write these sequences to two separate files. From Bio import SeqIO. FullAmyloid beta A4 protein.
In the case of DNA the nucleotides are represented using their one letter acronyms. Continue if line. For keyvalue in genesitems.
A T C and G. For line in file_one. In terminal write the following command python fasta_fixpy.
Count count 1. For seq_record in SeqIOparseopen file_in moder fasta. Running this script will generate a FASTA file with the header information and entire sequence as a single line.
With open file_out w as f_out. Heres an example of FASTA text. SeqIOwrite record fastq fastq The record is a SeqRecord object fastq is the file handle and fastq.
This is a basic example of Bioinformatics problem. You just need to write the name and sequence to a fasta format name1. 27082014 Id like feedback on my function to convert FASTA data to a dictionary in Python.
The typical way to write an ASCII fastq is done as follows. Ofile openmy_fastatxt w for i in rangelenlist_seq. Parsing a fasta file would be as simple as this.
I have been trying to read the fasta sequence but somehow it is always skipping the last sequence. Could you please help me what am I missing here. It can be used for both nucleotide and protein sequences.
FASTA files are used to store sequence data. FullAlzheimer disease amyloid A4 protein homolog. 15082020 Following is an example of how to read a FASTA file.
FASTA file format is a commonly used DNA and protein sequence file format. Check your folder and open the output FASTA file. Import sys fasta test with open testsfasta as file_one.
06022019 Using our testfasta file that we created count the number of records. Biopython - read and write a fasta file. From Bio import SeqIO for record in SeqIOparsefilenamefasta fasta.
List_namei n. The FASTA file format. If youre working with Python.
This very tutorial is about how to read Fasta file using python scripting. This is the fourth Tutorial about basics of Bioinformatics in python3 in this tutorial I go through the process of writing FASTA files in python 3. Most of the time that I work with a FASTA file in Python I load all the sequences into a dictionary where the sequences id is the key and the sequence is the value and I use the dictionary to run my analysis.
Count 0 for seq_record in SeqIO. It already contains this parsing functionality and a whole lot more. 27032019 To run the script go into fasta-fix folder using terminal.
With openinputfiler as f. Example FASTA output from my Github. List_seqi n do not forget to close it ofileclose.
Id rather avoid a separate system call. How do you write a gz or bgz fastq file using Biopython.

Length Filter A Fasta Subterranaut

Python Protein Sequence File Fasta Split Programmer Sought

Quantitative Extraction Specified Sequence From The Fasta File Python Script Programmer Sought

Rename Changing File Extension In Python Stack Overflow
Biopython Tutorial And Cookbook
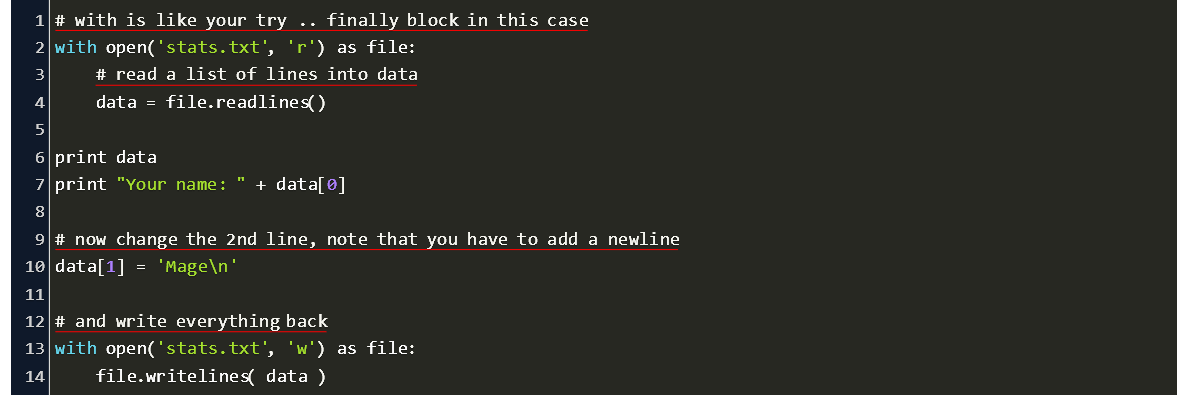
How To Edit A Specific Line In Text File In Python Code Example
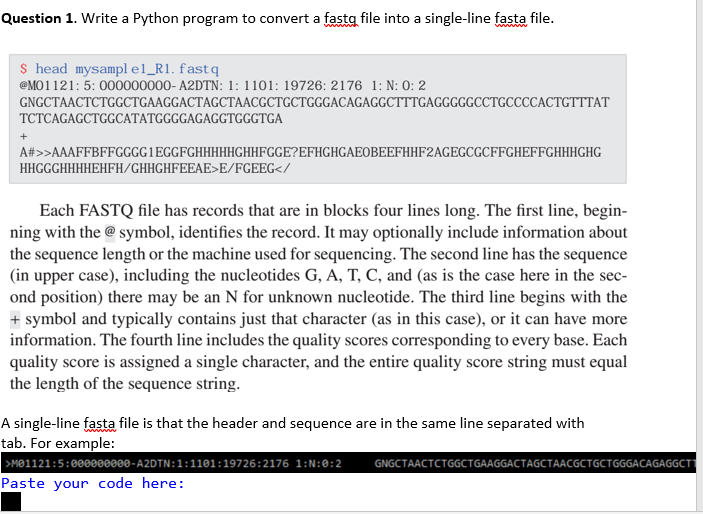
Question 1 Write A Python Program To Convert A Fasta Chegg Com
How To Write A Function Python 3 8 To Find Fasta Entry For Dna Sequence

Problem 1 Creating A Fasta Class Fasta File Reader Chegg Com

0 comments:
Post a Comment